
Redoxoma Highlights, by Francisco R. M. Laurindo
Corresponding author e-mail: francisco.laurindo@hc.fm.usp.br
As scientists, likely we would love to be what we think, but for practical purposes we are what we write. Wouldn't that be interesting to have a big picture of what words we have been writing in Redoxoma? Obviously, computing this is no easy task, but at least looking at the titles of our publications could give us a hint of which are our favorite subjects. Reporting word statistics would certainly be boring, however word clouds can provide quantitative and unbiased yet understable estimates. After all, if it is true that a picture is worth a thousand words, one should be able to learn something from a picture made of words.
With this idea in mind, I took the titles of all papers published in our Cepid-Redoxoma (from our site) an grouped them in 3 datasets: 2013 to 2015, 2016 to 2017 and 2018 to mid-2019. The aim was to contribute to those collections of amusing and [not-that-] useless trivia.
Obviously the datasets are not quantitatively equal, but doing some experimentation with distinct modes of organization did not yield relevant alterations in the overall results. Word Clouds were constructed using WordItOut software, organizing the word size by frequency. Results are shown in the figures below. As expected, a few bits of information can be derived from them. Here I bring a few interesting highlights and invite our readers to add additional observations in our site !
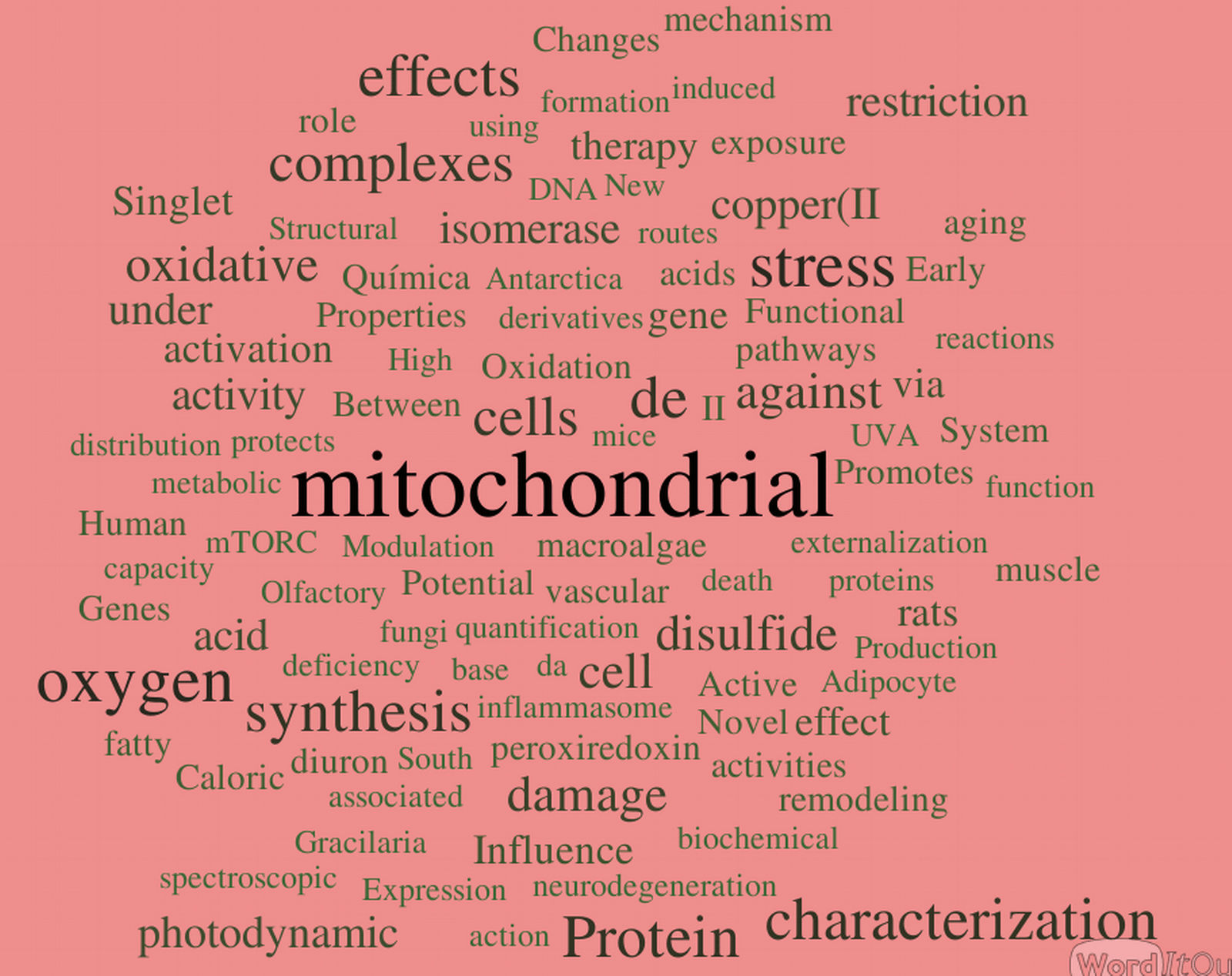
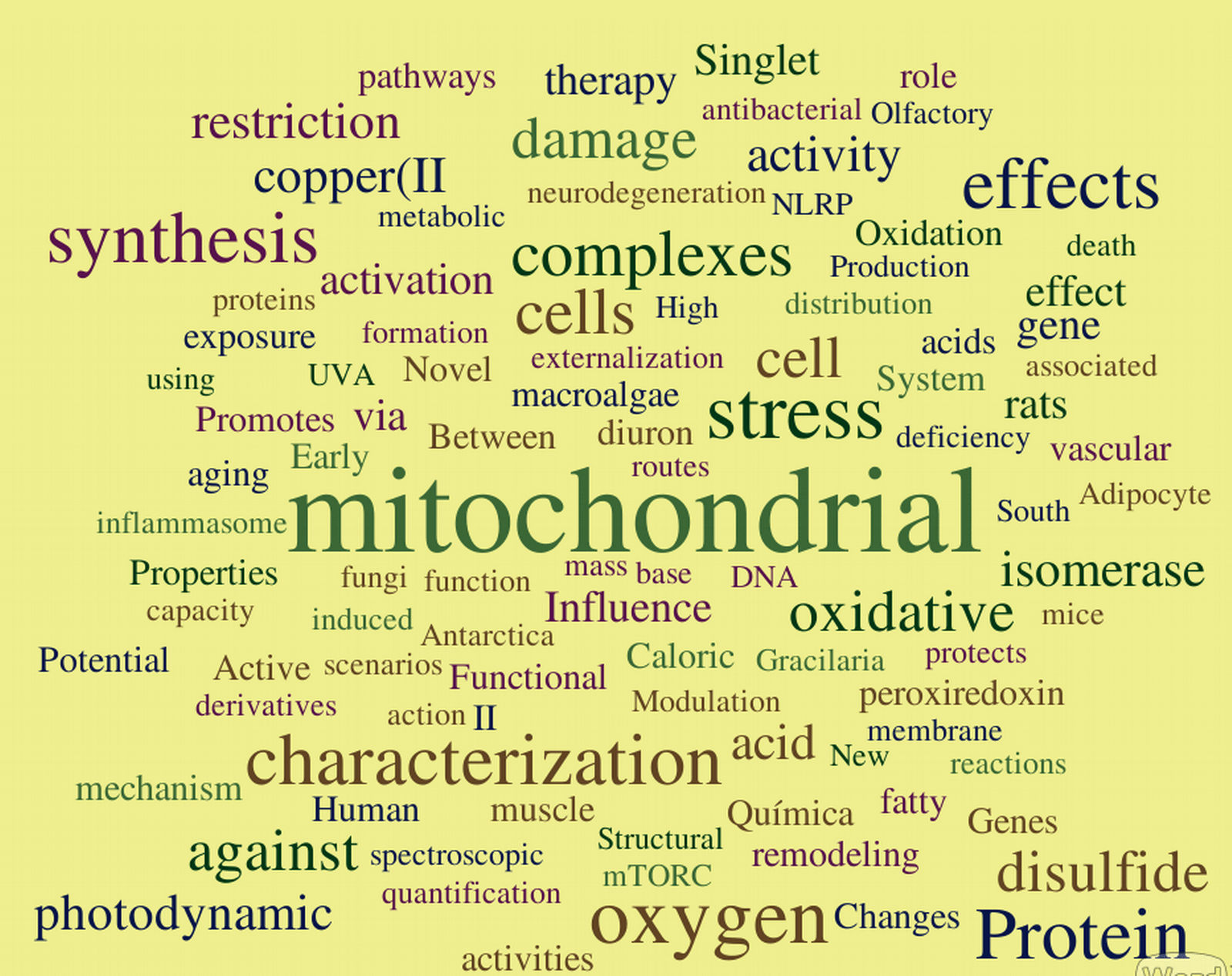
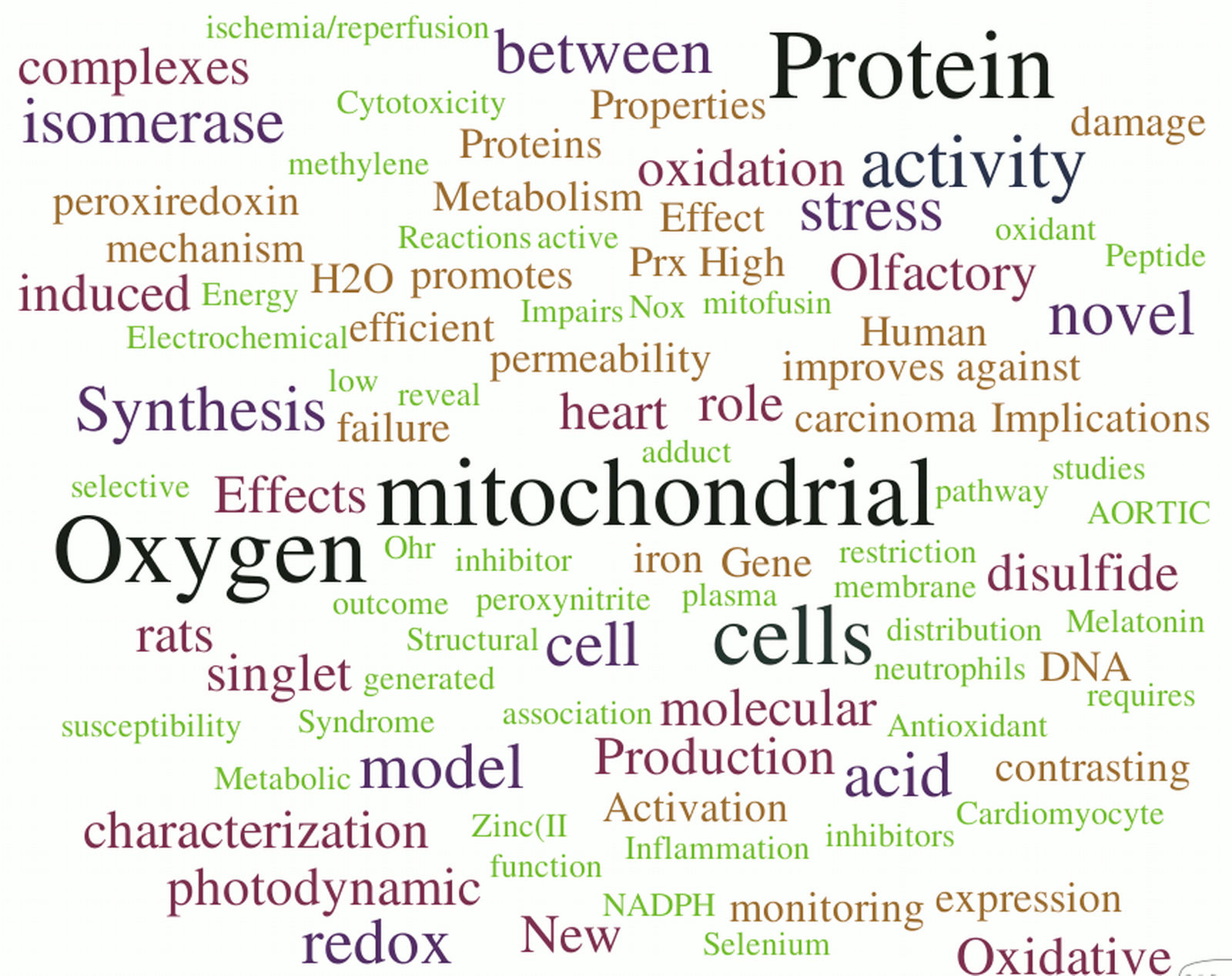
- Our main keywords are quite conserved across the years. This indicates a consistency of the central interests and research tracks, which I see as a faithful portrait of Redoxoma: focus on solid questions away from fashionisms. The risk of a lack of innovative and refreshing ideas, I believe, is readily dismissed by a closer exam of the published articles.
- Clearly, "mitochondrial" is the outstanding keyword in all datasets (we all know who is responsible for that...) no matter how we group the data. Recently, however, this remarkable organelle has been challenged by "oxygen"- quite a fair game I would say, given the true respectability of the basic element our work is based on.
- The words indicate we are definitely a protein rather than a gene group. Among the proteins, thiol proteins are steadily prominent: peroxiredoxins (Prx), isomerase (which derives from the PDI works, no personal bias please), Ohr, etc.
- Oxidant-related wording (oxidant, oxidative, disulfide, peroxynitrite) quite clearly predominates over reductant-related wording. This suggests that oxidation still dominates the collective uncounscious of redox investigators. Maybe it would be a good time to focus on reducing pathways?
- Some words are notable for their absence or paucity: the top candidate to me is "antioxidant".
- It was reassuring that manicheist jargon, such as "deleterious", "protective", "defense", "damaging", "toxic", etc does not dominate our wording. This reflects a state-of-the-art approach to free radical-dependent biology and medicine, in which thoughts are driven towards mechanisms rather than good-or-bad classifications.
- The words tell us we definitely seem to focus more on death rather than survival. And stress of course. Not surprising.
- I was surprised that "singlet" consistently outperformed "peroxide", "superoxide" and related words. Similarly, "photodynamic", not a usual word in redox biomedicine, was consistently high accross all datasets. Congratulations to all those involved.
- Copper was the most popular metal in the two earliest datasets, but now it has been surpassed by iron, with zinc and selenium as emerging runnerups. An interesting competition to watch.
- Not unexpectedly, there is good competition between methods/chemistry-related keywords and application/physiology ones. Will [spectroscopy, electrochemical, mass, quantification, activities, synthesis, reactions, adduct, etc...] outcompete [metabolic, vascular, aortic, heart, neurodegeneration, olfactory, caloric, etc...] ? The jury is out, but my personal veredict is that we should switch the word "competition" to "balance".
- "Human" stands nearly at the same rank as "rats" , but both are largely surpassed by "cell(s)". Also not surprising for a mechanistic group.
- The most recent dataset shows a wellcome emergence/increase of some interesting terms: "structural", "molecular", "model" and "mechanism". Hopefully we will keep writing more about these.
- In a context for the ugliest words, my vote would stay with "characterization" and "effects", which are description-related terms.
- The words tell us that for sure we kept doing many "novel" and "new" things at Redoxoma.
It is important to say that these exercises have several intrinsic biases from the scientific standpoint and should not be taken as a picture of the science we do at Redoxoma. Hopefully, however, these diagrams will give us some material for fueling our conversations and maybe some thoughts. And some fun, of course.
Francisco R. M. Laurindo, Editor in Chief of Redoxoma Newsletter at Heart Institute (InCor),
University of São Paulo Medical School, Brazil
Add new comment